Publications
Publications in peer reviewed journals
New methods to unravel rhizosphere processes
2016 - Trends Plant Sci., 3: 243-55
Abstract:
Root-triggered processes (growth, uptake and release of solutes) vary in space and time, and interact with heterogeneous soil microenvironments that provide habitats for (micro)biota on various scales. Despite tremendous progress in method development in the past decades, finding a suitable experimental set-up to investigate processes occurring at the dynamic conjunction of biosphere, hydrosphere, and pedosphere in the close vicinity of active plant roots still represents a major challenge. We discuss recent methodological developments in rhizosphere research with a focus on imaging techniques. We further review established concepts that have been updated with novel techniques, highlighting the need for combinatorial approaches to disentangle rhizosphere processes on relevant scales.
Fenton's reagent for the rapid and efficient isolation of microplastics from wastewater.
2016 - Chem. Commun. (Camb.), 2: 372-375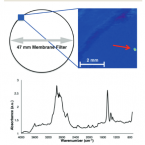
Abstract:
Fenton's reagent was used to isolate microplastics from organic-rich wastewater. The catalytic reaction did not affect microplastic chemistry or size, enabling its use as a pre-treatment method for focal plane array-based micro-FT-IR imaging. Compared with previously described microplastic treatment methods, Fenton's reagent offers a considerable reduction in sample preparation times.
IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies.
2016 - Sci Rep, 33721
Abstract:
The SRA (Sequence Read Archive) serves as primary depository for massive amounts of Next Generation Sequencing data, and currently host over 100,000 16S rRNA gene amplicon-based microbial profiles from various host habitats and environments. This number is increasing rapidly and there is a dire need for approaches to utilize this pool of knowledge. Here we created IMNGS (Integrated Microbial Next Generation Sequencing), an innovative platform that uniformly and systematically screens for and processes all prokaryotic 16S rRNA gene amplicon datasets available in SRA and uses them to build sample-specific sequence databases and OTU-based profiles. Via a web interface, this integrative sequence resource can easily be queried by users. We show examples of how the approach allows testing the ecological importance of specific microorganisms in different hosts or ecosystems, and performing targeted diversity studies for selected taxonomic groups. The platform also offers a complete workflow for de novo analysis of users' own raw 16S rRNA gene amplicon datasets for the sake of comparison with existing data. IMNGS can be accessed at www.imngs.org.
Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses
2016 - Nature, 537: 689–693
Abstract:
Ocean microbes drive biogeochemical cycling on a global scale1. However, this cycling is constrained by viruses that affect community composition, metabolic activity, and evolutionary trajectories2, 3. Owing to challenges with the sampling and cultivation of viruses, genome-level viral diversity remains poorly described and grossly understudied, with less than 1% of observed surface-ocean viruses known4. Here we assemble complete genomes and large genomic fragments from both surface- and deep-ocean viruses sampled during the Tara Oceans and Malaspina research expeditions5, 6, and analyse the resulting ‘global ocean virome’ dataset to present a global map of abundant, double-stranded DNA viruses complete with genomic and ecological contexts. A total of 15,222 epipelagic and mesopelagic viral populations were identified, comprising 867 viral clusters (defined as approximately genus-level groups7, 8). This roughly triples the number of known ocean viral populations4 and doubles the number of candidate bacterial and archaeal virus genera8, providing a near-complete sampling of epipelagic communities at both the population and viral-cluster level. We found that 38 of the 867 viral clusters were locally or globally abundant, together accounting for nearly half of the viral populations in any global ocean virome sample. While two-thirds of these clusters represent newly described viruses lacking any cultivated representative, most could be computationally linked to dominant, ecologically relevant microbial hosts. Moreover, we identified 243 viral-encoded auxiliary metabolic genes, of which only 95 were previously known. Deeper analyses of four of these auxiliary metabolic genes (dsrC, soxYZ, P-II (also known as glnB) and amoC) revealed that abundant viruses may directly manipulate sulfur and nitrogen cycling throughout the epipelagic ocean. This viral catalog and functional analyses provide a necessary foundation for the meaningful integration of viruses into ecosystem models where they act as key players in nutrient cycling and trophic networks.
Bacterial nutrient foraging in a mouse model of enteral nutrient deprivation: Insight into the gut origin of sepsis
2016 - Am J Physiol Gastrointest Liver Physiol, 311: G734-G743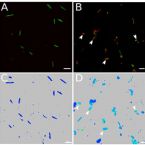
Abstract:
Total parenteral nutrition (TPN) leads to a shift in small intestinal microbiota with a characteristic dominance of Proteobacteria. This study examined how metabolomic changes within the small bowel support an altered microbial community in enterally deprived mice.
C57BL/6 mice were given TPN or enteral chow. Metabolomic analysis of jejunal contents was performed by liquid chromatography/mass spectrometry (LC/MS). In some experiments, leucine in TPN was partly substituted with (13)C-leucine. Additionally, jejunal contents from TPN dependent and enterally fed mice were gavaged into germ-free mice to reveal if the TPN phenotype was transferrable.
Small bowel contents of TPN mice maintained an amino acid composition similar to that of the TPN solution. Mass spectrometry analysis of small bowel contents of TPN dependent mice showed increased concentration of (13)C compared to fed mice receiving saline enriched with (13)C-leucine. (13)C-leucine added to the serosal side of Ussing chambers showed rapid permeation across TPN-dependent jejunum, suggesting increased transmucosal passage. Single-cell analysis by fluorescence in situ hybridization (FISH) - NanoSIMS demonstrated uptake of (13)C-leucine by TPN-associated bacteria, with preferential uptake by Enterobacteriaceae. Gavage of small bowel effluent from TPN mice into germ-free, fed mice resulted in a trend toward the pro-inflammatory TPN-phenotype with loss of epithelial barrier function.
TPN-dependence leads to increased permeation of TPN-derived nutrients into the small intestinal lumen, where they are predominately utilized by Enterobacteriaceae. The altered metabolomic composition of the intestinal lumen during TPN promotes dysbiosis.Single cell stable isotope probing in microbiology using Raman microspectroscopy
2016 - Curr Opin Biotechnol, 41:34-42
Abstract:
Microbial communities are essential for most ecosystem processes and interact in highly complex ways with virtually all eukaryotes. Thus, a detailed understanding of the function of such communities is a fundamental prerequisite for microbial ecologists, applied microbiologists and microbiome researchers. Using single cell Raman microspectroscopy, biochemical fingerprints of individual microbial cells can be obtained in a fast and nondestructive manner. If combined with stable isotope probing (SIP), Raman spectroscopy can directly reveal functions of single microorganisms in their natural habitat. This review provides an update on various SIP-approaches suitable for combination with different Raman scattering techniques and illustrates how single cell Raman SIP can be directly combined with the omics-centric analysis pipelines generally applied to investigate microbial communities.
Behavior of platinum(iv) complexes in models of tumor hypoxia: cytotoxicity, compound distribution and accumulation
2016 - Metallomics, 4: 422-33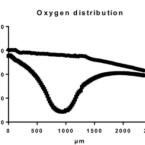
Abstract:
Hypoxia in solid tumors remains a challenge for conventional cancer therapeutics. As a source for resistance, metastasis development and drug bioprocessing, it influences treatment results and disease outcome. Bioreductive platinum(iv) prodrugs might be advantageous over conventional metal-based therapeutics, as biotransformation in a reductive milieu, such as under hypoxia, is required for drug activation. This study deals with a two-step screening of experimental platinum(iv) prodrugs with different rates of reduction and lipophilicity with the aim of identifying the most appropriate compounds for further investigations. In the first step, the cytotoxicity of all compounds was compared in hypoxic multicellular spheroids and monolayer culture using a set of cancer cell lines with different sensitivities to platinum(ii) compounds. Secondly, two selected compounds were tested in hypoxic xenografts in SCID mouse models in comparison to satraplatin, and, additionally, (LA)-ICP-MS-based accumulation and distribution studies were performed for these compounds in hypoxic spheroids and xenografts. Our findings suggest that, while cellular uptake and cytotoxicity strongly correlate with lipophilicity, cytotoxicity under hypoxia compared to non-hypoxic conditions and antitumor activity of platinum(iv) prodrugs are dependent on their rate of reduction.
Multi-scale imaging of anticancer platinum(IV) compounds in murine tumor and kidney
2016 - Chemical Science, 7: 3052-3061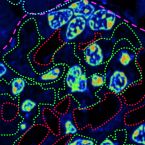
Abstract:
Nano-scale secondary ion mass spectrometry (NanoSIMS) enables trace element and isotope analyses with high spatial resolution. This unique capability has recently been exploited in several studies analyzing the subcellular distribution of Au and Pt anticancer compounds. However, these studies were restricted to cell culture systems. To explore the applicability to the in vivo setting, we developed a combined imaging approach consisting of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS), NanoSIMS and transmission electron microscopy (TEM) suitable for multi-scale detection of the platinum distribution in tissues. Applying this approach to murine tumor and kidney samples upon administration of selected platinum(IV) anticancer prodrugs revealed uneven platinum distributions on both the organ and subcellular scales. Spatial platinum accumulation patterns by LA-ICP-MS were quantitatively assessed in histologically heterogeneous organs (e.g., higher platinum accumulation in kidney cortex than in medulla) and used to select regions of interest for subcellular scale imaging with NanoSIMS. These analyses revealed cytoplasmic sulfur-rich organelles to accumulate platinum in both kidney and malignant cells. Those in the tumor were subsequently identified as organelles of lysosomal origin, demonstrating the potential of the combinatorial approach for investigating therapeutically relevant drug concentrations on a submicrometer scale.
Diversity analysis of sulfite- and sulfate-reducing microorganisms by multiplex dsrA and dsrB amplicon sequencing using new primers and mock community-optimized bioinformatics
2016 - Environ Microbiol, 18: 2994-3009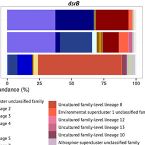
Abstract:
Genes encoding dissimilatory sulfite reductase (DsrAB) are commonly used as diagnostic markers in ecological studies of sulfite- and sulfate-reducing microorganisms. Here, we developed new high-coverage primer sets for generation of reductive bacterial-type dsrA and dsrB PCR products for highly parallel amplicon sequencing and a bioinformatics workflow for processing and taxonomic classification of short dsrA and dsrB reads. We employed two diverse mock communities that consisted of 45 or 90 known dsrAB sequences derived from environmental clones to precisely evaluate the performance of individual steps of our amplicon sequencing approach on the Illumina MiSeq platform. Although PCR cycle number, gene-specific primer mismatches, and stringent filtering for high-quality sequences had notable effects on the observed dsrA and dsrB community structures, recovery of most mock community sequences was generally proportional to their relative input abundances. Successful dsrA and dsrB diversity analysis in selected environmental samples further proved that the multiplex amplicon sequencing approach is adequate for monitoring spatial distribution and temporal abundance dynamics of dsrAB-containing microorganisms. While tested for reductive bacterial-type dsrAB, this method is readily applicable for oxidative-type dsrAB of sulfur-oxidizing bacteria and also provides guidance for processing short amplicon reads of other functional genes.
probeBase - an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016
2016 - Nucleic Acids Res, 44: D586-D589
Abstract:
probeBase http://www.probebase.net is a manually maintained and curated database of rRNA-targeted oligonucleotide probes and primers. Contextual information and multiple options for evaluating in silico hybridization performance against the most recent rRNA sequence databases are provided for each oligonucleotide entry, which makes probeBase an important and frequently used resource for microbiology research and diagnostics. Here we present a major update of probeBase, which was last featured in the NAR Database Issue 2007. This update describes a complete remodeling of the database architecture and environment to accommodate computationally efficient access. Improved search functions, sequence match tools, and data output now extend the opportunities for finding suitable hierarchical probe sets that target an organism or taxon at different taxonomic levels. To facilitate the identification of complementary probe sets for organisms represented by short rRNA sequence reads generated by amplicon sequencing or metagenomic analysis with next generation sequencing technologies such as Illumina and IonTorrent, we introduce a novel tool that recovers surrogate near full-length rRNA sequences for short query sequences and finds matching oligonucleotides in probeBase.