Publications
Publications in peer reviewed journals
New methods to unravel rhizosphere processes
2016 - Trends Plant Sci., 3: 243-55
Abstract:
Root-triggered processes (growth, uptake and release of solutes) vary in space and time, and interact with heterogeneous soil microenvironments that provide habitats for (micro)biota on various scales. Despite tremendous progress in method development in the past decades, finding a suitable experimental set-up to investigate processes occurring at the dynamic conjunction of biosphere, hydrosphere, and pedosphere in the close vicinity of active plant roots still represents a major challenge. We discuss recent methodological developments in rhizosphere research with a focus on imaging techniques. We further review established concepts that have been updated with novel techniques, highlighting the need for combinatorial approaches to disentangle rhizosphere processes on relevant scales.
Ecology and Fisheries: Dark Carbon on Your Dinner Plate.
2016 - Curr. Biol., 24: R1277-R1279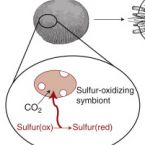
Abstract:
Chemosynthetic primary production by symbiotic microbes powers entire ecosystems in the remote deep sea. New research shows that in shallow waters chemosynthetic symbioses can contribute substantially to a vital economic resource - lobster fisheries in the Caribbean Sea.
Genome-guided design of a novel defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium
2016 - Nature Microbiol, 2: 16215
Abstract:
Protection against enteric infections, also termed colonization resistance, results from mutualistic interactions of the host and its indigenous microbes. The gut microbiota of humans and mice is highly diverse and it is therefore challenging to assign specific properties to its individual members. Here, we have used a collection of murine bacterial strains and a modular design approach to create a minimal bacterial community that, once established in germ-free mice, provided colonization resistance against the human enteric pathogen Salmonella enterica serovar Typhimurium (S. Tm). Initially, a community of 12 strains, termed Oligo-Mouse Microbiota (Oligo-MM12), representing members of the major bacterial phyla in the murine gut, was selected. This community was stable over consecutive mouse generations and provided colonization resistance against S. Tm infection, albeit not to the degree of a conventional complex microbiota. Comparative (meta)genome analyses identified functions represented in a conventional microbiome but absent from the Oligo-MM12. By genome-informed design, we created an improved version of the Oligo-MM community harbouring three facultative anaerobic bacteria from the Mouse Intestinal Bacterial Collection (miBC) that provided conventional-like colonization resistance. In conclusion, we have established a highly versatile experimental system that showed efficacy in an enteric infection model. Thus, in combination with exhaustive bacterial strain collections and systems-based approaches, genomeguided design can be used to generate insights into microbe–microbe and microbe–host interactions for the investigation of ecological and disease-relevant mechanisms in the intestine.
Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation.
2016 - Nat Microbiol, 2: 16195
Abstract:
Chemosynthetic symbioses are partnerships between invertebrate animals and chemosynthetic bacteria. The latter are the primary producers, providing most of the organic carbon needed for the animal host's nutrition. We sequenced genomes of the chemosynthetic symbionts from the lucinid bivalve Loripes lucinalis and the stilbonematid nematode Laxus oneistus. The symbionts of both host species encoded nitrogen fixation genes. This is remarkable as no marine chemosynthetic symbiont was previously known to be capable of nitrogen fixation. We detected nitrogenase expression by the symbionts of lucinid clams at the transcriptomic and proteomic level. Mean stable nitrogen isotope values of Loripes lucinalis were within the range expected for fixed atmospheric nitrogen, further suggesting active nitrogen fixation by the symbionts. The ability to fix nitrogen may be widespread among chemosynthetic symbioses in oligotrophic habitats, where nitrogen availability often limits primary productivity.
Free-living amoebae and their associated bacteria in Austrian cooling towers: a 1-year routine screening
2016 - Parasitol Res, 115: 3365-74
Abstract:
Free-living amoebae (FLA) are widely spread in the environment and known to cause rare but often serious infections. Besides this, FLA may serve as vehicles for bacterial pathogens. In particular, Legionella pneumophila is known to replicate within FLA thereby also gaining enhanced infectivity. Cooling towers have been the source of outbreaks of Legionnaires' disease in the past and are thus usually screened for legionellae on a routine basis, not considering, however, FLA and their vehicle function. The aim of this study was to incorporate a screening system for host amoebae into a Legionella routine screening. A new real-time PCR-based screening system for various groups of FLA was established. Three cooling towers were screened every 2 weeks over the period of 1 year for FLA and Legionella spp., by culture and molecular methods in parallel. Altogether, 83.3 % of the cooling tower samples were positive for FLA, Acanthamoeba being the dominating genus. Interestingly, 69.7 % of the cooling tower samples were not suitable for the standard Legionella screening due to their high organic burden. In the remaining samples, positivity for Legionella spp. was 25 % by culture, but overall positivity was 50 % by molecular methods. Several amoebal isolates revealed intracellular bacteria.
Microbial dinitrogen fixation in coral holobionts exposed to thermal stress and bleaching.
2016 - Environ. Microbiol., 8: 2620-33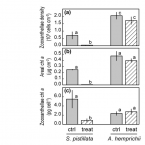
Abstract:
Coral holobionts (i.e., coral-algal-prokaryote symbioses) exhibit dissimilar thermal sensitivities that may determine which coral species will adapt to global warming. Nonetheless, studies simultaneously investigating the effects of warming on all holobiont members are lacking. Here we show that exposure to increased temperature affects key physiological traits of all members (herein: animal host, zooxanthellae and diazotrophs) of both Stylophora pistillata and Acropora hemprichii during and after thermal stress. S. pistillata experienced severe loss of zooxanthellae (i.e., bleaching) with no net photosynthesis at the end of the experiment. Conversely, A. hemprichii was more resilient to thermal stress. Exposure to increased temperature (+ 6°C) resulted in a drastic increase in daylight dinitrogen (N2 ) fixation, particularly in A. hemprichii (threefold compared with controls). After the temperature was reduced again to in situ levels, diazotrophs exhibited a reversed diel pattern of activity, with increased N2 fixation rates recorded only in the dark, particularly in bleached S. pistillata (twofold compared to controls). Concurrently, both animal hosts, but particularly bleached S. pistillata, reduced both organic matter release and heterotrophic feeding on picoplankton. Our findings indicate that physiological plasticity by coral-associated diazotrophs may play an important role in determining the response of coral holobionts to ocean warming.
Bacterial nutrient foraging in a mouse model of enteral nutrient deprivation: Insight into the gut origin of sepsis
2016 - Am J Physiol Gastrointest Liver Physiol, 311: G734-G743
Abstract:
Total parenteral nutrition (TPN) leads to a shift in small intestinal microbiota with a characteristic dominance of Proteobacteria. This study examined how metabolomic changes within the small bowel support an altered microbial community in enterally deprived mice.
C57BL/6 mice were given TPN or enteral chow. Metabolomic analysis of jejunal contents was performed by liquid chromatography/mass spectrometry (LC/MS). In some experiments, leucine in TPN was partly substituted with (13)C-leucine. Additionally, jejunal contents from TPN dependent and enterally fed mice were gavaged into germ-free mice to reveal if the TPN phenotype was transferrable.
Small bowel contents of TPN mice maintained an amino acid composition similar to that of the TPN solution. Mass spectrometry analysis of small bowel contents of TPN dependent mice showed increased concentration of (13)C compared to fed mice receiving saline enriched with (13)C-leucine. (13)C-leucine added to the serosal side of Ussing chambers showed rapid permeation across TPN-dependent jejunum, suggesting increased transmucosal passage. Single-cell analysis by fluorescence in situ hybridization (FISH) - NanoSIMS demonstrated uptake of (13)C-leucine by TPN-associated bacteria, with preferential uptake by Enterobacteriaceae. Gavage of small bowel effluent from TPN mice into germ-free, fed mice resulted in a trend toward the pro-inflammatory TPN-phenotype with loss of epithelial barrier function.
TPN-dependence leads to increased permeation of TPN-derived nutrients into the small intestinal lumen, where they are predominately utilized by Enterobacteriaceae. The altered metabolomic composition of the intestinal lumen during TPN promotes dysbiosis.Budget of primary production and dinitrogen fixation in a highly seasonal Red Sea coral reef
2016 - Ecosystems, 5: 771-785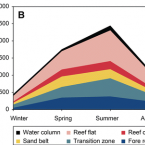
Abstract:
Biological dinitrogen (N2) fixation (diazotrophy, BNF) relieves marine primary producers of nitrogen (N) limitation in a large part of the world oceans. N concentrations are particularly low in tropical regions where coral reefs are located, and N is therefore a key limiting nutrient for these productive ecosystems. In this context, the importance of diazotrophy for reef productivity is still not resolved, with studies up to now lacking organismal and seasonal resolution. Here, we present a budget of gross primary production (GPP) and BNF for a highly seasonal Red Sea fringing reef, based on ecophysiological and benthic cover measurements combined with geospatial analyses. Benthic GPP varied from 215 to 262 mmol C m−2 reef d−1, with hard corals making the largest contribution (41–76%). Diazotrophy was omnipresent in space and time, and benthic BNF varied from 0.16 to 0.92 mmol N m−2 reef d−1. Planktonic GPP and BNF rates were respectively approximately 60- and 20-fold lower than those of the benthos, emphasizing the importance of the benthic compartment in reef biogeochemical cycling. BNF showed higher sensitivity to seasonality than GPP, implying greater climatic control on reef BNF. Up to about 20% of net reef primary production could be supported by BNF during summer, suggesting a strong biogeochemical coupling between diazotrophy and the reef carbon cycle.
Biophysical and Population Genetic Models Predict the Presence of “Phantom” Stepping Stones Connecting Mid-Atlantic Ridge Vent Ecosystems
2016 - Current Biology, 26: 1 - 11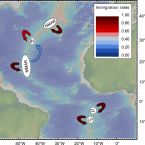
Abstract:
Deep-sea hydrothermal vents are patchily distributed ecosystems inhabited by specialized animal populations that are textbook meta-populations. Many vent-associated species have free-swimming, dispersive larvae that can establish connections between remote populations. However, connectivity patterns among hydrothermal vents are still poorly understood because the deep sea is undersampled, the molecular tools used to date are of limited resolution, and larval dispersal is difficult to measure directly. A better knowledge of connectivity is urgently needed to develop sound environmental management plans for deep-sea mining. Here, we investigated larval dispersal and contemporary connectivity of ecologically important vent mussels (Bathymodiolus spp.) from the Mid-Atlantic Ridge by using high-resolution ocean modeling and population genetic methods. Even when assuming a long pelagic larval duration, our physical model of larval drift suggested that arrival at localities more than 150 km from the source site is unlikely and that dispersal between populations requires intermediate habitats (“phantom” stepping stones). Dispersal patterns showed strong spatiotemporal variability, making predictions of population connectivity challenging. The assumption that mussel populations are only connected via additional stepping stones was supported by contemporary migration rates based on neutral genetic markers. Analyses of population structure confirmed the presence of two southern and two hybridizing northern mussel lineages that exhibited a substantial, though incomplete, genetic differentiation. Our study provides insights into how vent animals can disperse between widely separated vent habitats and shows that recolonization of perturbed vent sites will be subject to chance events, unless connectivity is explicitly considered in the selection of conservation areas.
A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria.
2016 - Trends Microbiol., 9: 699-712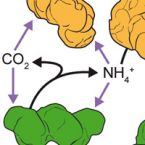
Abstract:
Nitrite-oxidizing bacteria (NOB) catalyze the second step of nitrification, nitrite oxidation to nitrate, which is an important process of the biogeochemical nitrogen cycle. NOB were traditionally perceived as physiologically restricted organisms and were less intensively studied than other nitrogen-cycling microorganisms. This picture is in contrast to new discoveries of an unexpected high diversity of mostly uncultured NOB and a great physiological versatility, which includes complex microbe-microbe interactions and lifestyles outside the nitrogen cycle. Most surprisingly, close relatives to NOB perform complete nitrification (ammonia oxidation to nitrate) and this finding will have far-reaching implications for nitrification research. We review recent work that has changed our perspective on NOB and provides a new basis for future studies on these enigmatic organisms.
Gypsum amendment to rice paddy soil stimulated bacteria involved in sulfur cycling but largely preserved the phylogenetic composition of the total bacterial community
2016 - Environ Microbiol Rep, 8: 413-23
Abstract:
Rice paddies are indispensable for human food supply but emit large amounts of the greenhouse gas methane. Sulfur cycling occurs at high rates in these water-submerged soils and controls methane production, an effect that is increased by sulfate-containing fertilizers or soil amendments. We grew rice plants until their late vegetative phase with and without gypsum (CaSO4 ·2H2 O) amendment and identified responsive bacteria by 16S rRNA gene amplicon sequencing. Gypsum amendment decreased methane emissions by up to 99% but had no major impact on the general phylogenetic composition of the bacterial community. It rather selectively stimulated or repressed a small number of 129 and 27 species-level operational taxonomic units (OTUs) (out of 1,883-2,287 observed) in the rhizosphere and bulk soil, respectively. Gypsum-stimulated OTUs were affiliated with several potential sulfate-reducing (Syntrophobacter, Desulfovibrio, unclassified Desulfobulbaceae, unclassified Desulfobacteraceae) and sulfur-oxidizing taxa (Thiobacillus, unclassified Rhodocyclaceae), while gypsum-repressed OTUs were dominated by aerobic methanotrophs (Methylococcaceae). Abundance correlation networks suggested that two abundant (>1%) OTUs (Desulfobulbaceae, Rhodocyclaceae) were central to the reductive and oxidative parts of the sulfur cycle.
Trophosome of the deep-sea tubeworm Riftia pachyptila inhibits bacterial growth
2016 - PLoS One, 11: e0146446
Abstract:
The giant tubeworm Riftia pachyptila lives in symbiosis with the chemoautotrophic gammaproteobacterium Cand. Endoriftia persephone. Symbionts are released back into the environment upon host death in high-pressure experiments, while microbial fouling is not involved in trophosome degradation. Therefore, we examined the antimicrobial effect of the tubeworm's trophosome and skin. The growth of all four tested Gram-positive, but only of one of the tested Gram-negative bacterial strains was inhibited by freshly fixed and degrading trophosome (incubated up to ten days at either warm or cold temperature), while no effect on Saccharomyces cerevisiae was observed. The skin did not show antimicrobial effects. A liquid chromatography-mass spectrometric analysis of the ethanol supernatant of fixed trophosomes lead to the tentative identification of the phospholipids 1-palmitoleyl-2-lyso-phosphatidylethanolamine, 2-palmitoleyl-1-lyso-phosphatidylethanolamine and the free fatty acids palmitoleic, palmitic and oleic acid, which are known to have an antimicrobial effect. As a result of tissue autolysis, the abundance of the free fatty acids increased with longer incubation time of trophosome samples. This correlated with an increasing growth inhibition of Bacillus subtilis and Listeria welshimeri, but not of the other bacterial strains. Therefore, the free fatty acids produced upon host degradation could be the cause of inhibition of at least these two bacterial strains.
Chlamydial seasonal dynamics and isolation of 'Candidatus Neptunochlamydia vexilliferae' from a Tyrrhenian coastal lake.
2016 - Environ. Microbiol., 8: 2405-17
Abstract:
The Chlamydiae are a phylum of obligate intracellular bacteria comprising important human and animal pathogens, yet their occurrence in the environment, their phylogenetic diversity and their host range has been largely underestimated. We investigated the seasonality of environmental chlamydiae in a Tyrrhenian coastal lake. By catalysed reporter deposition fluorescence in situ hybridization, we quantified the small planktonic cells and detected a peak in the abundance of environmental chlamydiae in early autumn with up to 5.9 × 10(4) cells ml(-1) . Super-resolution microscopy improved the visualization and quantification of these bacteria and enabled the detection of pleomorphic chlamydial cells in their protist host directly in an environmental sample. To isolate environmental chlamydiae together with their host, we applied a high-throughput limited dilution approach and successfully recovered a Vexillifera sp., strain harbouring chlamydiae (93% 16S rRNA sequence identity to Simkania negevensis), tentatively named 'Candidatus Neptunochlamydia vexilliferae'. Transmission electron microscopy in combination with fluorescence in situ hybridization was used to prove the intracellular location of these bacteria representing the first strain of marine chlamydiae stably maintained alongside with their host in a laboratory culture. Taken together, this study contributes to a better understanding of the distribution and diversity of environmental chlamydiae in previously neglected marine environments.
A Rickettsiales symbiont of amoebae with ancient features.
2016 - Environ. Microbiol., 8: 2326-42
Abstract:
The Rickettsiae comprise intracellular bacterial symbionts and pathogens infecting diverse eukaryotes. Here, we provide a detailed characterization of 'Candidatus Jidaibacter acanthamoeba', a rickettsial symbiont of Acanthamoeba. The bacterium establishes the infection in its amoeba host within 2 h where it replicates within vacuoles. Higher bacterial loads and accelerated spread of infection at elevated temperatures were observed. The infection had a negative impact on host growth rate, although no increased levels of host cell lysis were seen. Phylogenomic analysis identified this bacterium as member of the Midichloriaceae. Its 2.4 Mb genome represents the largest among Rickettsiales and is characterized by a moderate degree of pseudogenization and a high coding density. We found an unusually large number of genes encoding proteins with eukaryotic-like domains such as ankyrins, leucine-rich repeats and tetratricopeptide repeats, which likely function in host interaction. There are a total of three divergent, independently acquired type IV secretion systems, and 35 flagellar genes representing the most complete set found in an obligate intracellular Alphaproteobacterium. The deeply branching phylogenetic position of 'Candidatus Jidaibacter acanthamoeba' together with its ancient features place it closely to the rickettsial ancestor and helps to better understand the transition from a free-living to an intracellular lifestyle.